Systematic search and isolation of microbial secondary metabolites
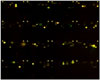
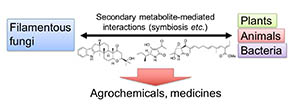
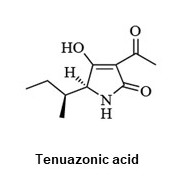
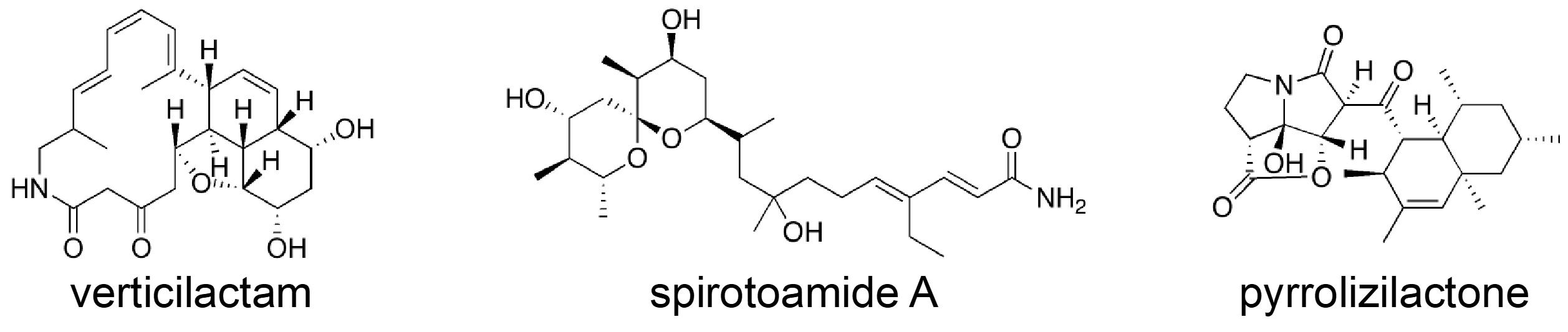
Microbes such as Streptomyces and fungi produce various structurally unique secondary metabolites with interesting biological activities. Such metabolites have been used for medicinal drugs, agrochemicals, and bioprobes to investigate biological functions in a chemical biology study. To search and isolate such important metabolites efficiently, we have constructed a microbial metabolite fraction library, which contains semi-purified extracts generated by basic chromatographic techniques. The library is analyzed by DAD-LC/MS to afford physicochemical information including retention time, UV and mass spectra of each metabolite within the fraction. Based on the information, we have constructed an original database named NPPlot (Natural Products Plot). It is a distribution map in which metabolites are appeared as dots in 2D area by retention time and molecular weight. Combination of the fraction library and NPPlot is a powerful tool for screening of structurally novel and interesting metabolites, and we have isolated various interesting metabolites by the system.






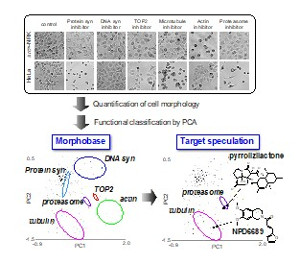
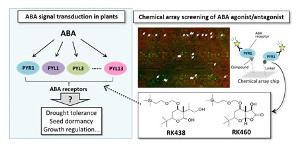
 In chemical genetics, the technology for high-throughput screening of a bioprobe regulating a protein function is very important. The identification of small-molecule bioprobes for a protein of interest can facilitate not only the functional analysis of the protein but also the development of clinical drugs.
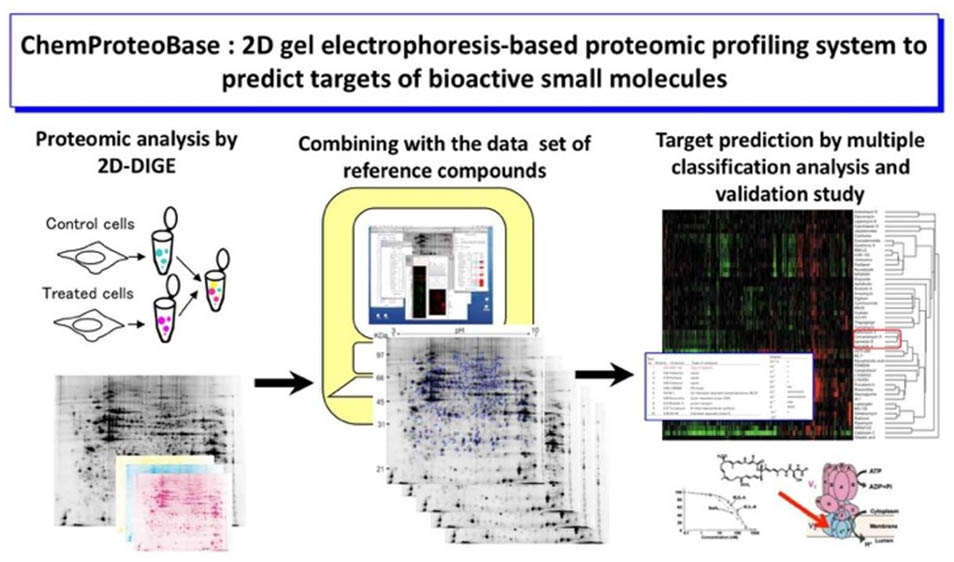
We have developed chemical array "NPDepoArray" for ultra-high throughput screening of bioprobes. The chemical array contains 2-3 thousands of small molecules immobilized with a unique photo-cross-linking approach in a functional-group-independent manner. Chemical array-based screens have enabled the discovery of small molecules that bind target proteins of interest.
In chemical genetics, the technology for high-throughput screening of a bioprobe regulating a protein function is very important. The identification of small-molecule bioprobes for a protein of interest can facilitate not only the functional analysis of the protein but also the development of clinical drugs.
We have developed chemical array "NPDepoArray" for ultra-high throughput screening of bioprobes. The chemical array contains 2-3 thousands of small molecules immobilized with a unique photo-cross-linking approach in a functional-group-independent manner. Chemical array-based screens have enabled the discovery of small molecules that bind target proteins of interest.