Drug target analysis based on proteomic analysis by 2D-DIGE.
Outline:
The exposure of the cells to bioactive small-molecules induces specific changes of the proteome. Using two-dimensional difference gel electrophoresis (2D-DIGE), proteomic analysis of the HeLa cells treated with a new compound is performed. Based on the expression data of ~300 spots, we compare well-characterized inhibitors and new compounds by hierarchical cluster analysis.
Compounds:Iejimalide A, BNS-22
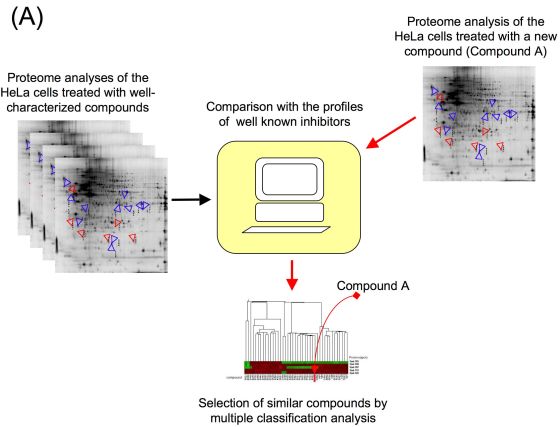
- Proteomic profiling system for drug target analysis using 2D-DIGE.(2)
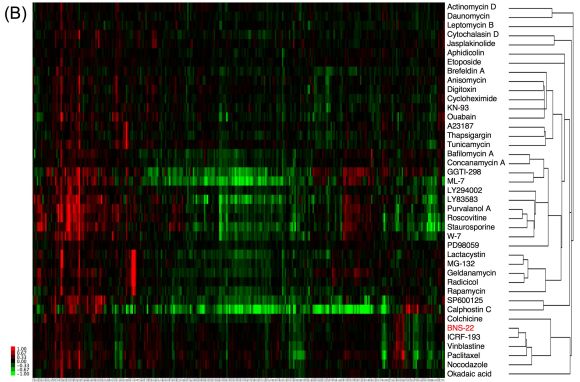
- Clustering of BNS-22 and well-known inhibitors based on the data of proteomic analysis using 2D-DIGE.
HeLa cells were treated with compound or with DMSO for 18 h were analyzed by 2D-DIGE and hierarchical cluster analysis was performed using the quantitative data of spots. The target of BNS-22 was estimated to Topo II by the system and inhibitory activity against Topo II was verified. This figure was modified based on the previously published data.(3)
References:
-
M. Muroi, S. Kazami, K. Noda, H. Kondo, H. Takayama, M. Kawatani, T. Usui, H. Osada.
Application of proteomic profiling based on 2D-DIGE for classification of compounds according to the mechanism of action,
Chem. Biol., 17, 460-70 (2010). -
K. Wierzba, M. Muroi, H. Osada.
Proteomics accelerating the identification of the target molecule of bioactive small molecules.
Curr. Opin. Chem.Biol., 15, 57-65 (2011). -
M. Kawatani, H. Takayama, M. Muroi, S. Kimura, T Maekawa, H. Osada.
Identification of a small-molecule inhibitor of DNA topoisomerase II by proteomic profiling,
Chem. Biol., 18, 743-751 (2011).


