Identification of target proteins using small molecule affinity matrix.
Outlines:
Affinity purification of small-molecule binding protein has been one of the most important techniques in forward chemical genetics. Once hit compound(s) which induce a specific phenotype in a cellular system are identified, the next step is to identify the molecular target.
Recently, we reported a coupling method that enables the introduction of a variety of small molecules onto glass slides through a photoaffinity reaction. By using this method, aryldiazirine groups covalently attached to glass slides are transformed upon irradiation with UV light into highly reactive carbenes, which in turn bind to or insert irreversibly into a proximal small molecule in a manner that is independent of the functional groups. This method, referred to as photo-cross-linking, has proven useful in the construction of small-molecule microarrays. It is expected that a similar chemical approach could be used to introduce small molecules onto affinity gel, such as onto agarose beads.
We have prepared PALC beads and demonstrated that photo-cross-linked small-molecule agarose beads can be used to identify and purify the binding protein. It was of note that complex small molecules such as FK506, rapamycin, and cyclosporin A could be introduced on the affinity matrix by using a uniform and straightforward procedure.
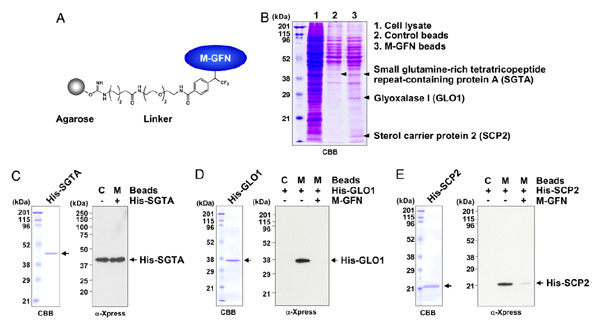 |
|
Identification of M-GFN-binding proteins. |
References:
-
N. Kanoh, K. Honda, S. Simizu, M. Muroi, H. Osada.
Photo-crosslinked small molecule affinity matrix for facilitating forward and reverse chemical genetics.
Angew. Chem. Int. Ed., 44, 3559-3562 (2005). -
M. Kawatani, H. Okumura, K. Honda, N. Kanoh, M. Muroi, N. Dohmae, M. Takami, M. Kitagawa, Y. Futamura, M. Imoto, H. Osada.
The Identification of an osteoclastogenesis inhibitor through the inhibition of glyoxalase I.
Proc. Natl. Acad. Sci. U S A., 105, 10691-11696 (2008).


